Advanced Gene Editing

Targeted genome editing has rapidly become a vital tool across the entire research continuum from early discovery to therapeutic application. By enabling scientists to selectively disrupt, recover, repress, or activate gene expression, modern gene editing methods allow for unprecedented exploration of genetic mechanisms governing biological processes. Application of genome engineering ranges from curing developmental disorders to developing disease-resistant crops.
Featured Categories
With decades of gene editing experience and an unsurpassed portfolio of CRISPR and ZFN technology patents, Sigma-Aldrich® Advanced Genomics delivers the highest quality reagents for your entire workflow, from early-stage discovery to translational research. Whether you are looking to knockout, knock-in, knockdown, or overexpress your targets, our comprehensive suite of CRISPR gene-editing tools and services will take your research Beyond the Bench.
Transfection reagents, protocols, and resources including Roche X-tremeGENE transfection reagents for DNA, siRNA, miRNA, and CRISPR/Cas9 gene editing components.
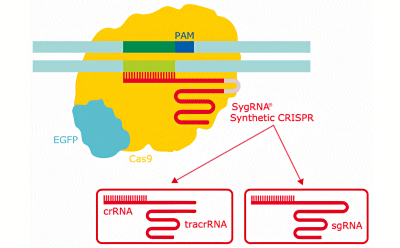
Schematic diagram of the CRISPR gene editing system showing the different types of synthetic gRNA for CRISPR.
CRISPR – Accurate, Efficient Gene Editing
Gene editing is a specific and targeted change to a DNA sequence and involves the addition, removal or modification of the DNA. The CRISPR-Cas system (evolved in microbes as a defense mechanism) is the basis for a class of gene-editing tools that are enabling advances from health and diagnostics to agriculture and energy. Using CRISPR, researchers have the power to target a specific gene, gene family, or even screen an entire genome.
Sigma-Aldrich® Advanced Genomics offers an industry leading selection of CRISPR-Cas9 proteins to meet your individual research needs. Choose from multiple Cas9 variants (wild-type, enhanced specificity, nickase, GFP-fused, catalytically inactive) and several formats (plasmid, lentivirus, lyophilized protein).
CRISPRi and CRISPRa – Powerful Gene Inhibition and Activation
CRISPRi (CRISPR interference) and CRISPRa (CRISPR activation) deliver highly efficient silencing and activation of genes, respectively, without altering the underlying DNA sequence. When employed in large scale LOF (loss-of-function) and GOF (gain-of-function) screens, researchers are able to identify unique, yet functionally related, gene pathways that are often missed with other methods.
Sigma-Aldrich® Advanced Genomics offers a complete suite of optimized CRISPRi and CRISPRa libraries for gene knockdown and overexpression experiments. Pooled CRISPRi and SAM CRISPRa lentiviral libraries are available off-the-shelf or customized to your specific needs.
Wondering if CRISPR is right for your project? Connect with a Sigma-Aldrich® Advanced Genomics Expert.
Visit our document search for data sheets, certificates and technical documentation.
Related Articles
- Get tips for handling lentiviruses, optimizing experiment setup, titering lentivirus particles, and selecting helpful products for transduction.
- Compare and contrast the features of a wide variety of guide RNA (gRNA) and Cas9 products for in vitro and in vivo CRISPR experiments.
- Automation ensures reproducible results in high-throughput assays like cell transfection, reducing manual handling variations.
- E. coli CRISPR/Cas-mediated Lambda-Red HR vector system enables gene editing via homology-directed repair.
- Techniques for long oligo synthesis and purification offer superior quality unmatched by other suppliers.
- See All (21)
Related Protocols
- Learn about CRISPR Cas9, what it is and how it works. CRISPR is a new, affordable genome editing tool enabling access to genome editing for all.
- Detailed procedure for how to perform a lentiviral transduction of MISSION shRNA lentiviral particles to achieve a stable long term silencing and phenotypic change.
- Combine guaranteed sgRNAs with our comprehensive range of CRISPR products and tools, including Cas9 and delivery reagents, for efficient genome modification with higher specificity.
- Guaranteed PURedit™ CRISPR synthetic gRNAs and Cas9 protein offer industry-leading on-site cutting and specificity
- ZFNs and CRISPR/Cas9 advance targeted genome editing, opening new research avenues.
- See All (7)
Find More Articles and Protocols
How Can We Help
In case of any questions, please submit a customer support request
or talk to our customer service team:
Email [email protected]
or call +1 (800) 244-1173
Additional Support
- CRISPR Use License Agreement
- Calculators & Apps
Web Toolbox - science research tools and resources for analytical chemistry, life science, chemical synthesis and materials science.
- Customer Support Request
Customer support including help with orders, products, accounts, and website technical issues.
- FAQ
Explore our Frequently Asked Questions for answers to commonly asked questions about our products and services.
To continue reading please sign in or create an account.
Don't Have An Account?















